paired end sequencing read length
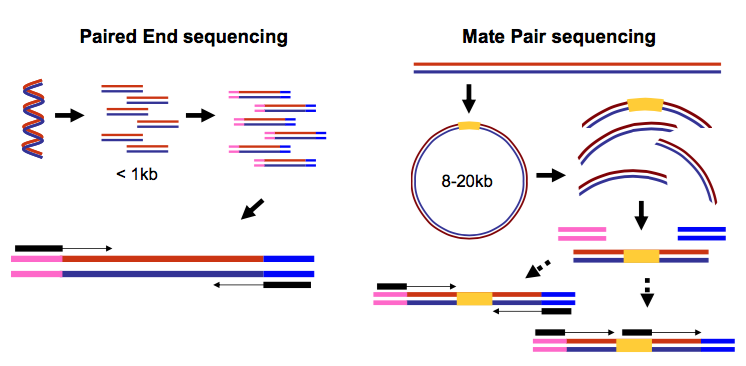
The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the. Simple workflow allows generation of unique ranges of insert sizes.
Ngs Bioinformatics Workshop 2 Ppt Download
To ensure sequencing quality of the Index Read do not exceed the supported read.

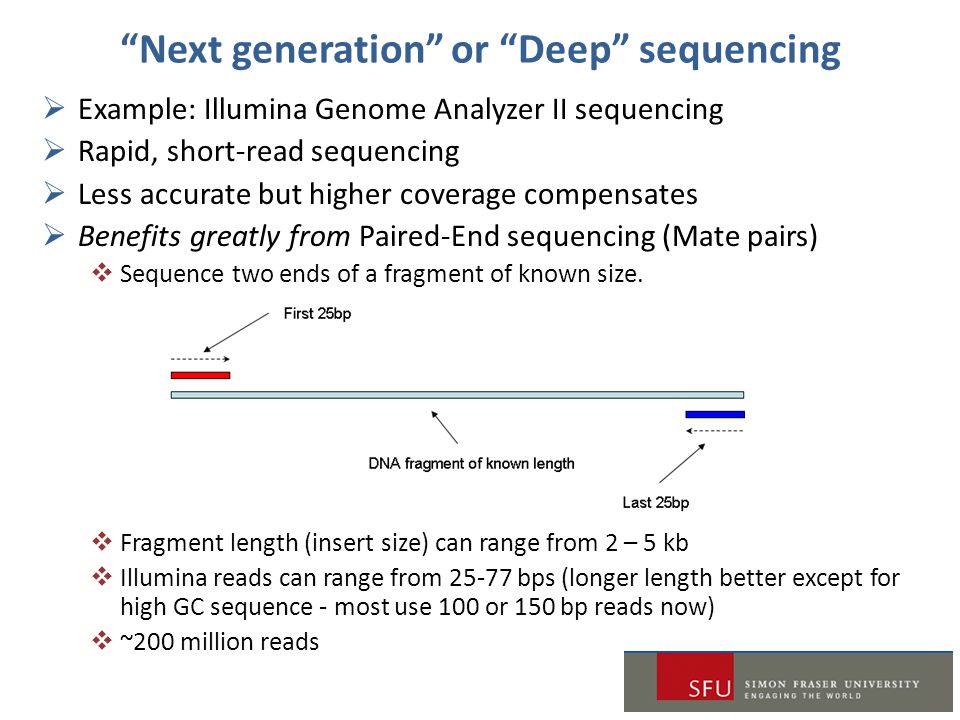
. Requires the same amount. A good choice for read length is closely tied to the insert size of the sequencing library ie how long the individual DNA fragments are that are sequenced. The pooled indexed libraries were loaded into the MiSeq Reagent kit V2 300 cycles Illumina and sequenced on the MiSeq platform Illumina in paired-end mode with a read.
An analysis by Whiteford et al. Whether you align 100bp paired reads or a. For example one read might consist of 50 base pairs 100 base pairs or more.
When using RNA-Seq to study gene expression read length is not a significant factor. On sequencing using unpaired reads shows that ultra-short reads theoretically allow whole genome re-sequencing and de novo assembly of only small. During sequencing it is possible to specify the number of base pairs that are read at a time.
Enter up to 20 characters or use manual mode if you need between 20 and 100 bp. We use an Illumina MiniSeq for our short-read sequencing runs. The paired-end short read lengths are always 2 x 150bp 300bp.
Application of sequencing to RNA analysis RNA-Seq whole transcriptome SAGE expression analysis novel organism mining splice variants Search in titles only Search in RNA. I think that the sequence of 2x250 bp came from miseq sequencing of V3-V4 region using 341F and 805R primers or so and it means that the insert is about 450 bp and about 430 bp. The Illumina paired-end sequencing technology can generate reads from both ends of target DNA fragments which can subsequently be merged to increase the.
What matters is read counts. The library prep protocols are designed to. This size depends on the library.

Assessing The Performance Of The Oxford Nanopore Technologies Minion Sciencedirect

Rna Extraction Method Read Length And Sequencing Layout Single End Versus Paired End Contribute Strongly To Variation Between Rna Seq Samples Rna Seq Blog

Paired End Vs Single End Sequencing Reads Youtube
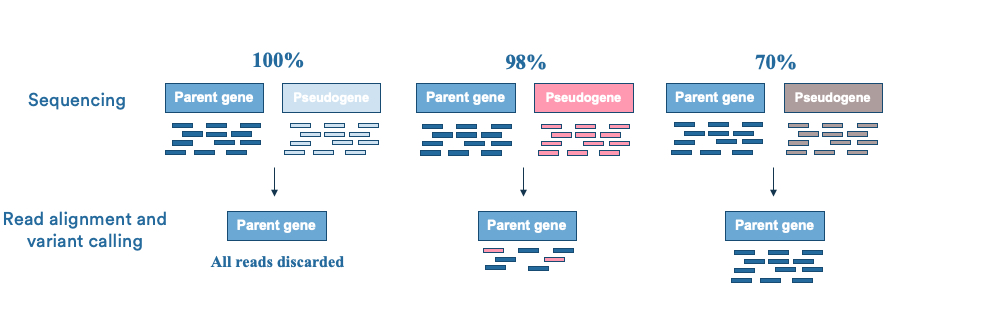
Blueprint Genetics Approach To Pseudogenes And Other Duplicated Genomic Regions Blueprint Genetics

Improving Nanopore Read Accuracy With The R2c2 Method Enables The Sequencing Of Highly Multiplexed Full Length Single Cell Cdna Pnas

Range Of Ngs Applications Rises Quickly
Short Read Sequencing Genomics Core Ecu
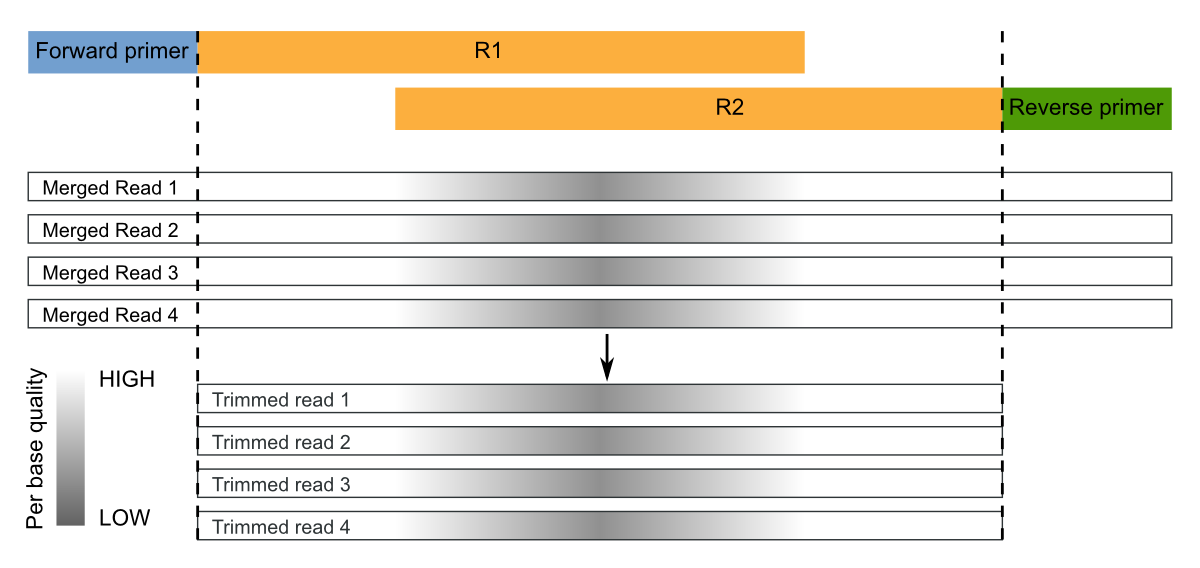
Paired End Sequencing 97 Otu Micca 1 7 0 Documentation
Illumina High Throughput Sequencing Dna Technologies Core
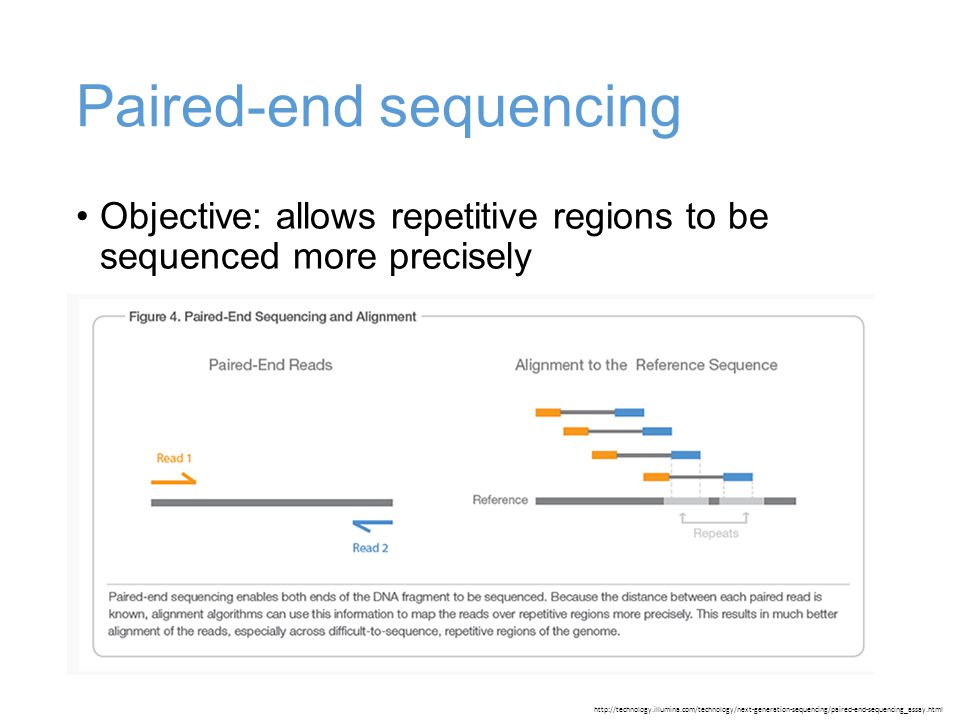
Mcb Lecture 9 Sept 23 14 Illumina Library Preparation De Novo Genome Assembly Ppt Download
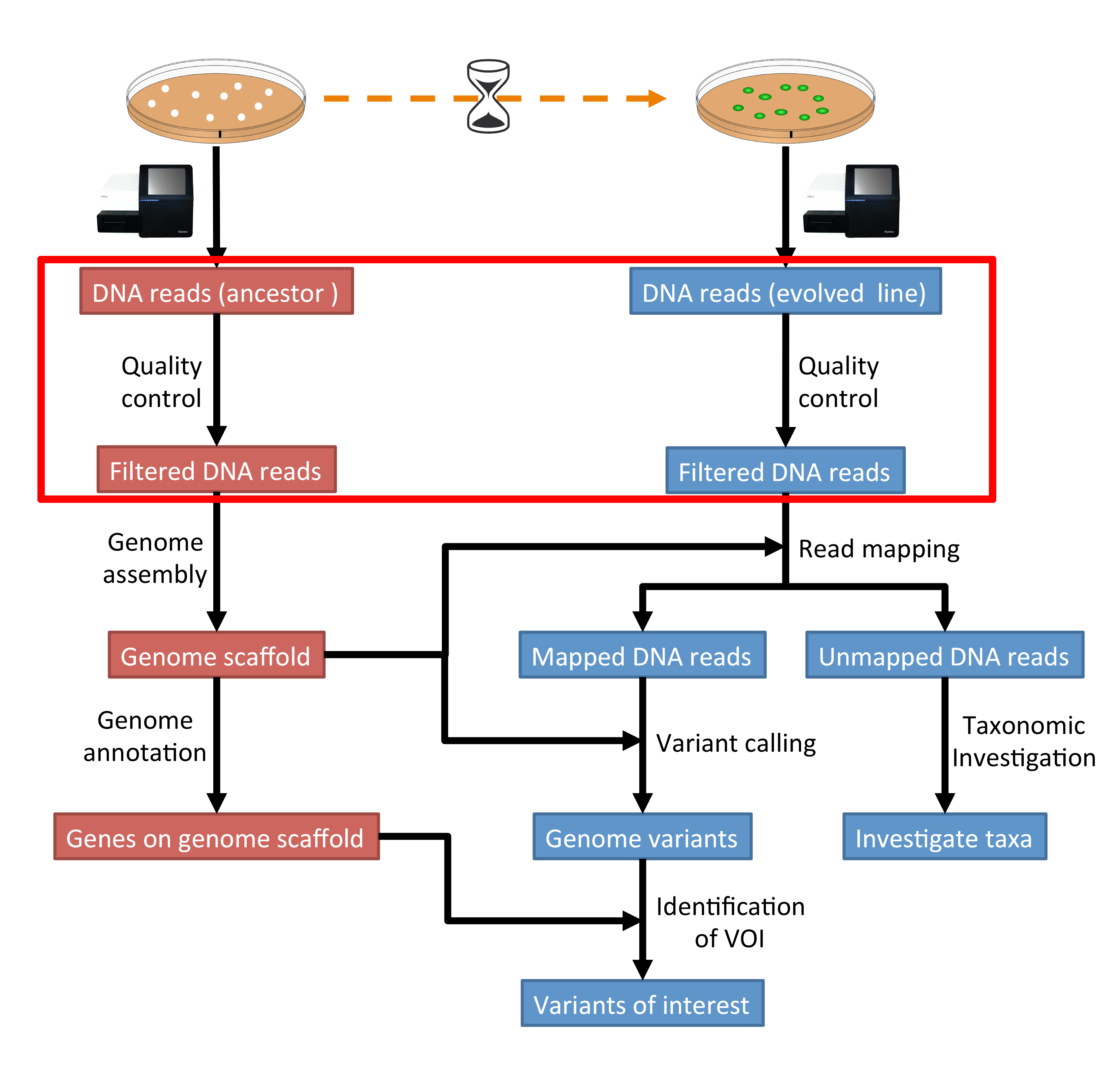
3 Quality Control Genomics Tutorial 2020 2 0 Documentation

Five Approaches To Detect Cnvs From Ngs Short Reads A Paired End Download Scientific Diagram

Rna Extraction Method Read Length And Sequencing Layout Single End Versus Paired End Contribute Strongly To Variatio Interactive Notebooks Method Sequencing
Local De Novo Assembly Of Rad Paired End Contigs Using Short Sequencing Reads Plos One
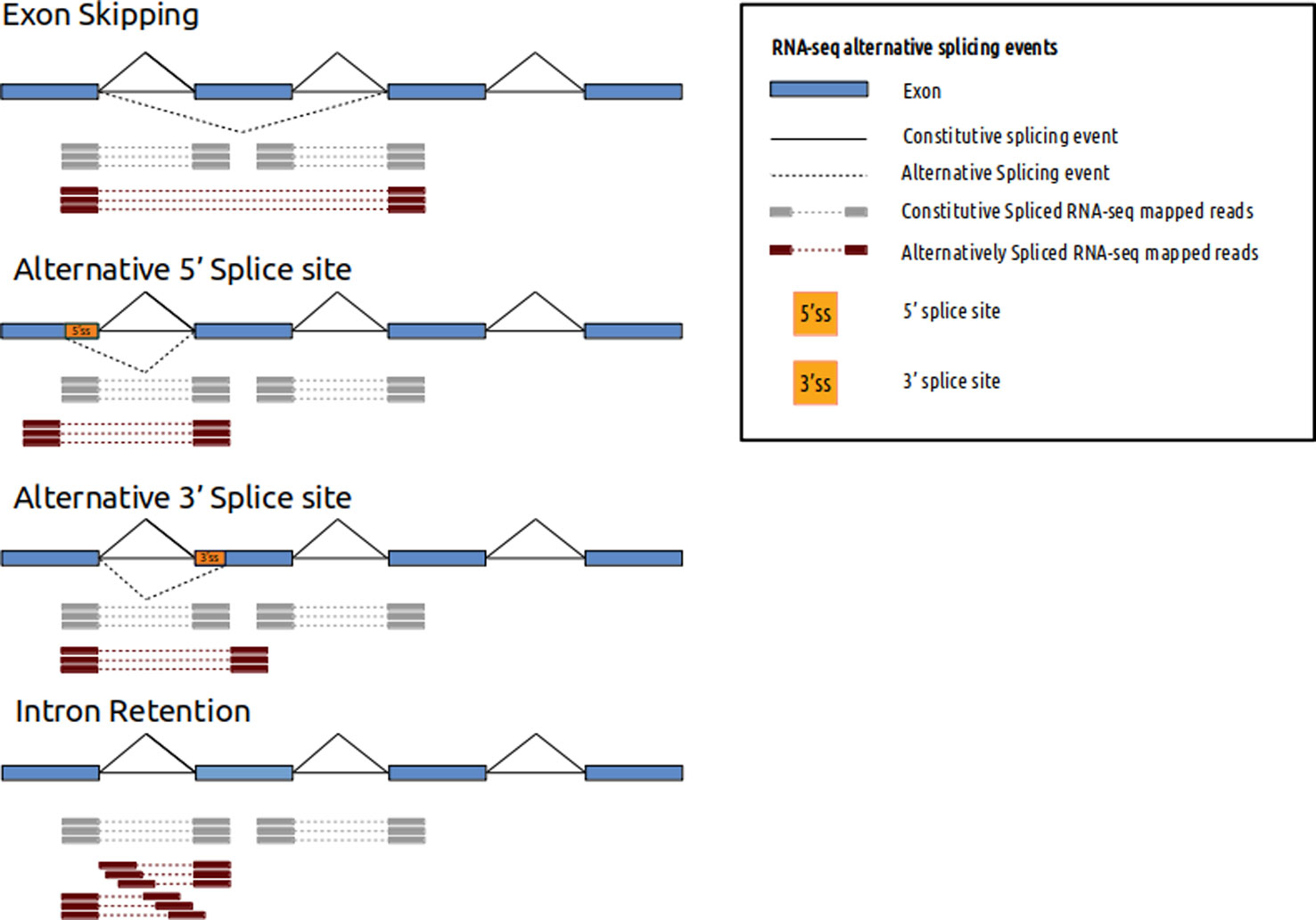
Single End Sequencing Versus Paired End
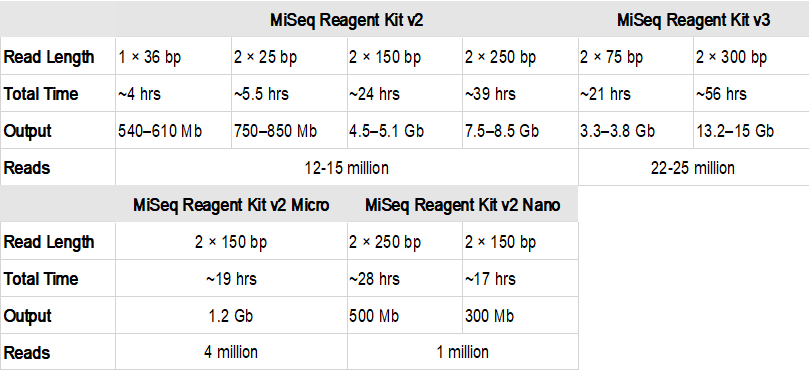
What 2 Indicates In 2 150 Bp 2 300 Bp Maximum Read Length From Illumina Website Http Www Illumina Com Systems Sequencing Platforms Html
A Comprehensive Evaluation Of Single End Sequencing Data Analyses For Environmental Microbiome Research Springerlink

Use Of Pairwise Linkage Information For Scaffolding A Paired End Download High Resolution Scientific Diagram